Accelerate Therapeutic Development of Genetic Diseases From SNP Functional Analysis to Drug Candidate
Introduction
Next generation sequencing has led to rapid expansion of our understanding of the complexity of human genetic variation. The complexity and consequences of single nucleotide polymorphisms (SNPs) and other human genetic variations1 have profound implications for future medicine. Sequence variations cataloged with association of diseases and benign phenotypes can both contribute to creating effective, precise, “personalized” therapies for unmet medical needs.
Moving from sequence analysis towards precision therapies requires
- identification of the specific genetic variant as the potential target
- functional validation of the consequences of the genetic variant
This has proven to be challenging and expensive to date. New tools such as antisense oligonucleotides (ASO), microRNAs (miRNA), CRISPR and targeted protein degradation (Protac) are now being employed (Figure 1) for rapid and precise dissection of biological pathways to speed up these steps. Multispan’s Functional Validation platform combines these approaches with our expertise in cell-based assays to enable the functional analysis of genetic variation. In this article, we discuss applications in the rare disease and broader therapeutic markets.
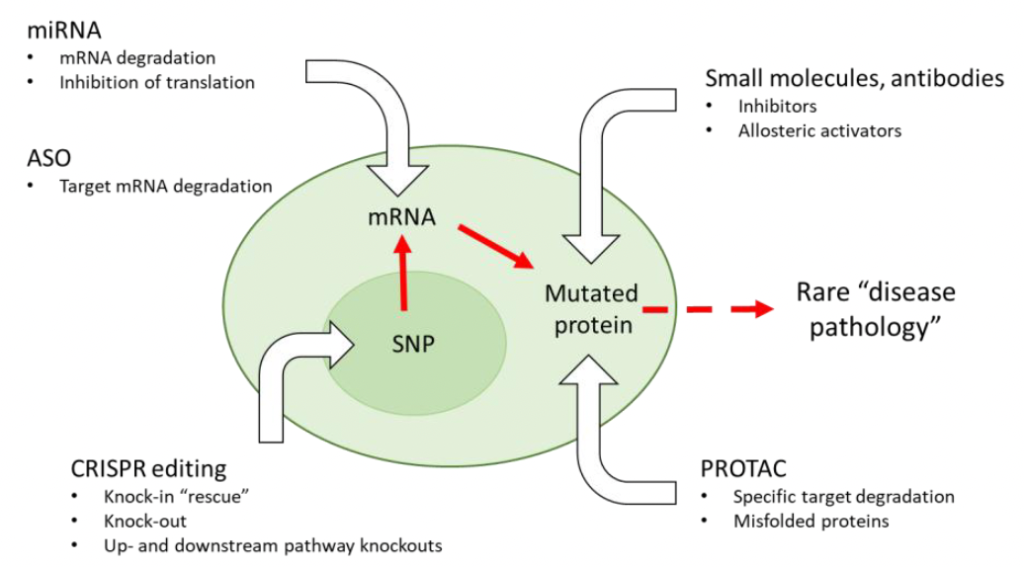
Figure 1 A coding region SNP encodes a protein that leads to rare disease pathology when expressed. CRISPR and other specific tools permit detailed functional dissection of the target and can be developed for therapeutic use
Functional validation of genetic variants associated with rare diseases
An estimated ~30M or 1 in 10 Americans suffer from a rare disease (classified as such impacting fewer two hundred thousand individuals1). Of 7000 catalogued rare diseases, up to 80% appear likely to be genetic in origin. FDA-approved therapies currently address fewer than 5% of these. The evolution of whole exome sequencing (WES) and other genotyping methods have expanded understanding of rare diseases in terms of sequence variation at the genome level2, 3, 4 However, for most genetic variants, insight into their functional role in the etiology of disease is lacking. This impedes the development of informative diagnostics and therapeutics. Validation of monogenic rare disease targets6 therefore requires 1) identification of sequence variants associated with disease and 2) the development of evidence that a specific genetic variant or SNP has a function in its pathophysiology. To meet this challenge, the cell-based platform created by Multispan enables the analysis of the impact of genetic variation on biological pathway activity in rare diseases.
Tools for therapeutics development targeting rare diseases
A major goal of rare disease research is to inform therapeutic intervention to cure or alleviate the effects of disease. Figure 1 outlines how Multispans’s approach feeds the development of high throughput screening (HTS) assays, animal models and other tools to support efforts to develop new therapeutics. In addition to classical therapeutic modalities (small molecules, antibodies) tools such as siRNA, CRISPR and Protac are beginning to be used to target rare diseases, either to downregulate activity or expression of a target, or in the case of CRISPR, to introduce a “corrected” copy of a mutated gene Collectively, these tools have the potential to extend the range of targets that may be considered as druggable. Since mutations that give rise to rare diseases may have either a gain (GOF)-or loss-of-function (LOF) effect on the target5,6, our expertise with this spectrum of therapeutic approaches paired with screening tools and animal models provides a flexible platform for targeting a specific disease. Indeed, Multispan has decades of accumulated expertise with targets impacted by both GOF and LOF mutations. For example, mutations in the GPCR rhodopsin can lead to the development of autosomal dominant retinitis pigmentosa7. A spectrum of GOF and LOF mutations also play a role in more than sixty other rare diseases currently associated with GPCRs7, 8.
The Multispan platform
Multispan’s platform takes the experimental approaches9 with our deep expertise in cell biology and assay development to address the following key questions about a potential target (Figure 2)
- What is known about the biological function of the target including its role in biological pathways and potential downstream effectors?
- What is known about the association between the target and pathophysiology of disease given that this may be indirect?
- Does genetic variation affect the primary structure, expression levels, cellular location, tissue expression of the target in disease-affected tissues?
- How does the biological activity of the normal and rare disease target compare in cellular assays?
We use targeted functional rescue to compare the cellular biology of variant and normal forms of the gene together with pathway dissection tools such as ASO, mRNA, CRISPR and Protac9,10. We combine data from these analyses, multi-omics assays and publicly available clinical, genetic and functional data analysis to provide a deep understanding of the effects of genetic variants on biological activity. These findings can identify potential biomarkers for further analysis, inform development of diagnostics as well as biochemical and/or cellular assays as well as tissue and animal models to inform downstream preclinical studies.
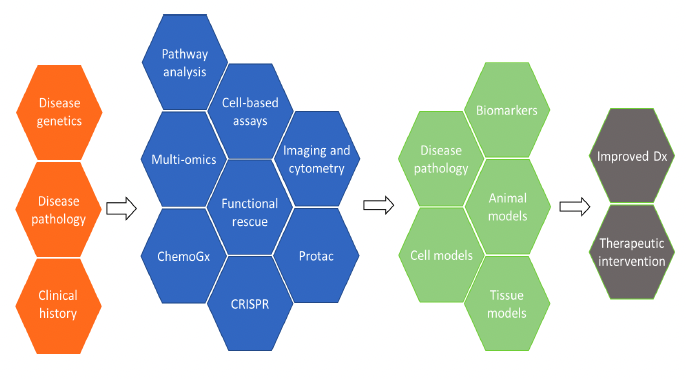
Figure 2 The Multispan Functional Validation Platform uses a combination of tools (blue) to study and validate the link between target and disease to create assays and models (green) leading to improved diagnostics and therapies
Precision Medicine and Pharmacogenomics
A second core application of the platform addresses the need for functional characterization of naturally occurring genetic variants of drug targets. These variations can give rise to variable responses to drugs, serious adverse reactions and pharmacoresistance11.
Analysis of the distribution of genetic variations in GPCRs provides a preliminary estimate of the scope of this problem for this important target class4,7,12. This work suggests that a “new, non-lethal, missense, germline mutation in a GPCR drug target arises in 1 of every ∼300 newborns.” This can impact “receptor conformation, dynamics, and signaling by affecting allosteric sites, effector selectivity sites, receptor stability, and oligomerization.” Multiple studies have documented the role of polymorphisms in the activity of other traditional drug targets such as ion channels and kinases13. As human sequence databases grow to include “sequencing from birth”, a richer picture of this variation and its potential significance will grow.
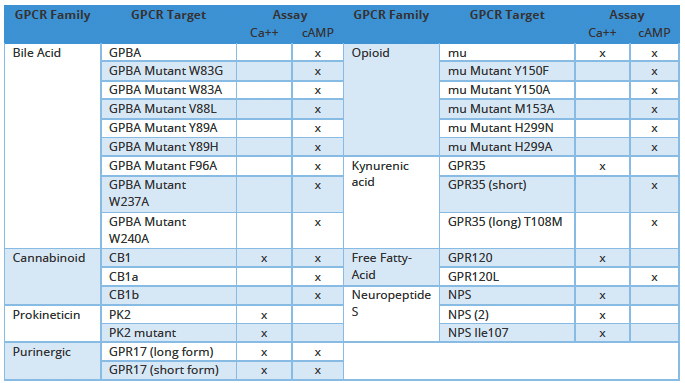
Our goal is therefore to support the assessment of the functional consequences of genetic variation during drug development. In support of this, we have previously described14 characterization of more than thirty cell lines expressing major and variant alleles well as of pharmaceutically relevant GPCR targets (Table 1). These include the bile acid receptor GPBAR1, mu-opioid receptors as well as tissue-specific isoforms of the CB1 cannabinoid receptor15.
For G-protein coupled receptor (GPCR) targets, our assays include calcium, cAMP, β-arrestin, pGRK2, IP-1, pERK, pNFκB, radioligand binding, GTPγS, chemotaxis, cytokine secretion, insulin secretion, cell proliferation, luciferase reporter and internalization. Multispan also has decades of accumulated experience in assay development for other target classes, including serine/threonine kinases, receptor tyrosine kinases, nuclear hormone receptors, transporters, ion channels, etc.
Table 1 Matched Wild-type vs Mutant and Canonical vs Variant GPCR Stable Cell Lines and Cell-based Assays
References
- RARE Disease Facts – Global Genes
- Boycott KM, Hartley T, Biesecker LG, et al. A diagnosis for all rare genetic diseases: the horizon and the next frontiers. Cell. 2019;177(1):32-37. PMID 30901545
- Exome Variant Server (washington.edu)
- Turner TN, Yi Q, Krumm N, et al. Denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 2017;45(D1):D804-D811. PMID 27907889
- Chen B, Altman RB. Opportunities for developing therapies for rare genetic diseases: focus on gain-of-function and allostery. Orphanet J Rare Dis. 2017;12(1):61. PMID: 28412959
- McInnes G, Sharo AG, Koleske ML, et al. Opportunities and challenges for the computational interpretation of rare variation in clinically important genes. Am J Hum Genet. 2021;108(4):535-548. PMID 33798442
- Schöneberg T, Liebscher I. Mutations in G protein-coupled receptors: mechanisms, pathophysiology and potential therapeutic approaches. Pharmacol Rev. 2021;73(1):89-119. PMID 33219147
- van der Velden WJC, Lindquist P, Madsen JS, et al. Molecular and in vivo phenotyping of missense variants of the human glucagon receptor. J Biol Chem. 2022;298(2):101413. doi:10.1016/j.jbc.2021.101413 PMID: 34801547
- Rodenburg RJ. The functional genomics laboratory: functional validation of genetic variants. J Inherit Metab Dis. 2018;41(3):297-307. PMID 29445992
- Yesbolatova A, Tominari Y, Kanemaki MT. Ligand-induced genetic degradation as a tool for target validation. Drug Discov Today Technol. 2019;31:91-98. PMID 31200864
- Barbarino JM, Whirl‐Carrillo M, Altman RB, Klein TE. PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip Rev Syst Biol Med. 2018;10(4):e1417.doi:10.1002/cpt.2349 PMID 29474005
- Hauser AS, Chavali S, Masuho I, et al. Pharmacogenomics of GPCR Drug Targets. Cell. 2018;172(1-2):41-54.e19. doi:10.1016/j.cell.2017.11.033 PMC5766829
- Raines R, McKnight I, White H, et al. Drug-targeted genomes: mutability of ion channels and GPCRs. Biomedicines. 2022;10(3):594. PMID 35327396
- Multispan GPCR Stable Cell Line Generation for Cell-Based Assay (multispaninc.com)
- González-Mariscal I, Krzysik-Walker SM, Doyle ME, et al. Human CB1 receptor isoforms, present in hepatocytes and β-cells, are involved in regulating metabolism. Sci Rep. 2016;6:33302. PMID 27641999

Recent Comments