Though 30% of all FDA-approved drugs target GPCRs, it wasn’t until 2018 that a monoclonal antibody therapy targeting a GPCR came to market. This is because the challenges to GPCR antibody development for both therapeutic uses and research are numerous. In this paper, we discuss the characteristics of GPCRs that make it so difficult to develop effective anti-GPCR antibodies. We then discuss Multispan’s customized MULTISCREENTM GPCR FACS binding and functional assays for antibody profiling.
G-protein coupled receptors (GPCRs) are essential for extracellular signal transduction mediating intracellular cell signaling processes. Their role in a variety of diseases, including cancer, chronic inflammatory conditions, neurological disorders, and infection, underscores their clinical relevance as attractive therapeutic targets and the importance of continued research. In fact, more than 30% of all FDA-approved drugs target GPCRs [1]. However, the development of monoclonal antibodies that target GPCRs for both R&D and therapeutic purposes is impeded by characteristics common to these receptors. This explains why, despite all the interest in targeting GPCRs, it wasn’t until 2018 that the first monoclonal antibody targeting a GPCR, erenumab, was approved by the FDA [2].
More than 800 human GPCRs have been identified [3]. GPCRs have 7-transmembrane regions with an extracellular N-terminus and intracellular C-terminus. The intracellular C-terminus interacts with heterotrimeric G-proteins. Representing the largest class of receptors, they play critical roles in physiological processes (e.g., immune responses, neurotransmission, olfaction) and disease pathogenesis. GPCR ligands are as diverse as the receptors themselves, including lipids, ions, nucleic acids, other proteins, and even photons.
There are extensive challenges to GPCR antibody development. First, due to the lipophilic environment in which GPCRs exist, it is very difficult to express recombinant GPCR antigens in a soluble form for antibody production. Second, the limited exposed area of these receptors provides few available epitopes and there exists extensive antibody cross-reactivity among receptors. Additionally, GPCRs have multiple active conformations. Finally, there are few efficient GPCR antibody screening tools.
The problems of expression and solubility
GPCRs have proven difficult to express at high levels using either cell-based or cell-free techniques. Functional GPCRs have been expressed in bacterial, yeast, insect, and mammalian cell systems with varying efficiencies. Cell-free synthesis methods have also been used, though these tend to be more expensive [4,5]. One such approach utilizes a bilayer-dialysis method [5].
To purify the receptors for downstream antibody generation, GPCRs expressed in cell-based systems must be extracted from the membrane. This is often accomplished using detergents. However, the transmembrane regions of GPCRs exist in a lipophilic environment. This feature makes it exceedingly difficult to solubilize recombinant GPCR antigens in their native form in hydrophilic conditions. The detergent-solubilized form and the natively folded protein can be quite dissimilar. Solubilized GPCRs are also prone to degradation. One engineering approach, called alanine scanning, has been used to identify and then selectively mutate the protein to stabilize the GPCR [6]. Another approach is purifying the GPCR in the presence of a suitable stabilizing ligand.
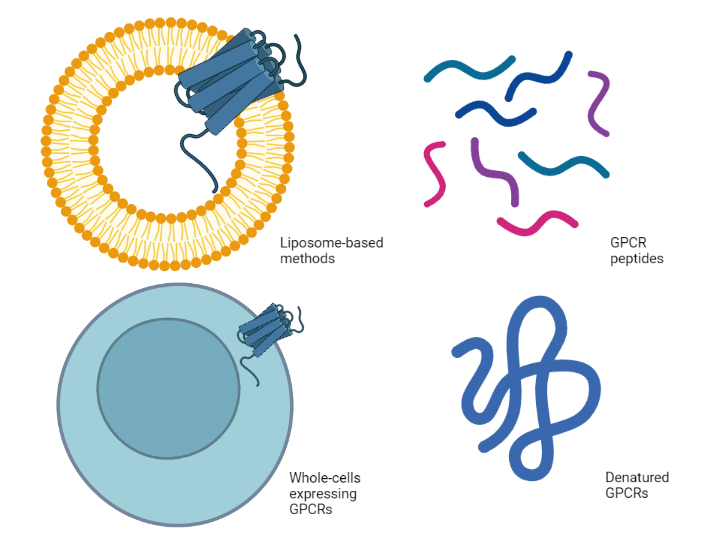
An alternative strategy is to abandon the isolation of intact, solubilized, properly folded GPCRs altogether. Synthetic peptides that represent extracellular portions of the receptor are often used, but these linear peptides cannot replicate the conformational structure of an intact GPCR. Similarly, denatured GPCRs have been used to successfully raise polyclonal antibodies against some human GPCRs [7]. Nevertheless, the concern regarding the limitations of using synthetic peptides and denatured proteins that cannot replicate the native form remain.
Perhaps unsurprisingly, liposome-based techniques have been developed to address the solubility issue. Mirzabekow et al. expressed CCR5 in canine thymocytes and then isolated the GPCR within its native membrane to retain the native form to use for selecting CCR5-specific antibodies [8]. A cell-free biotinylated liposome-based method was successfully used to identify antibodies against DRD1, GHSR, PTGER1, and T1R1 [5]. Whole cells expressing the GPCR have also been used to raise antibodies. Monoclonal antibodies against the human glucagon receptor (GCGR) were generated in humanized mice that were immunized with whole cells expressing human GCGR [9].
The problems of epitope availability and cross-reactivity
Much of a GPCR is embedded within the membrane. Antibodies raised against transmembrane portions of the protein would not be useful for therapeutic applications or many research techniques that involve intact cells or membranes (e.g., flow cytometry) as those regions would not be exposed. Given the small size of the extracellular region, epitopes in the exposed extracellular regions of the GPCR are limited.
Many GPCRs have similar sequences resulting in cross-reactivity of antibodies. Given the already limited epitope availability, this makes raising highly specific antibodies exceedingly difficult. Extensive screening is required to validate specificity (more on this later).
The problem of shape-shifting receptors
GPCRs can naturally take on many different conformational forms. Purification methods may add in several other conformational forms that are not relevant to any native state. This can result in generating antibodies against multiple forms. The shape-shifting nature of GPCRs layers on the challenges of the generation of functionally active monoclonal antibodies. Heterogeneity can also arise from other post-translational modifications such as N-glycosylation or receptor oligomerization.
The problem with antibody screening methods
Antibodies can be raised through the immunization of animals (e.g., mice, rabbits) with GPCR antigens in a variety of forms (e.g., protein, peptides, liposomes, whole cells). Alternatively, antibodies can be identified through high-throughput screening of antibody libraries – either from naïve or immune hosts or from synthetic libraries – using antibody display technologies. Of note, a recent study found that naïve libraries were just as effective as immune-sourced libraries [10]. Classically phage display was employed, but a few new display technologies exist today. The development of monoclonal antibodies using phage display or related technologies is integral to the use of monoclonal antibodies as therapeutics where mouse-derived antibodies have limited utility [11].
Phage display led the way to the first-ever FDA-approved human antibody therapy, Humira, in 2002 [12]. However, phage display is not without its limitations and the development of new screening technologies has proven difficult. Display technologies are limited by their libraries. Phage display libraries are robust, but this
can result in the generation of antibodies that would never arise in vivo. Simultaneously, some antigen-specific antibodies may not be present.
With the rise of new single-cell multi-omics technologies over the last few years, there is much promise in the use of single B cell technologies for antigen discovery [13].
Commercial antibodies
Millions of commercial antibodies for research purposes are available from hundreds of companies. Estimates for the percentage of these that are both target-specific and sensitive enough to detect endogenously expressed proteins vary widely, but some startling estimates predict that less than half are reliable [14].
How are commercial antibodies validated prior to going on the market? Not nearly as extensively as one might predict. Often validation is not repeated for subsequent lots and knockout cells are only now becoming more commonly used for validation. Cross-reactivity is rarely tested on any robust panel. Validation is often limited to visualizing a band via western blot at the predicted size, without necessary controls (such as knockout or null lines), and without validating for any other research techniques (e.g., flow cytometry, immunohistochemistry, immunoprecipitation).
Efforts to standardize validation and evaluate commercially available antibodies are gaining traction. Antibodypedia (https://www.antibodypedia.com/) is an open-access database of validated antibodies maintained by the Human Protein Atlas. Similarly, Antibodies-Online (https://www.antibodies-online.com/) and The Antibody Registry (https://antibodyregistry.org/) provide validation information for a wide range of antibodies.
MULTISCREENTM antibody profiling
For antibodies targeting GPCRs, sufficient validation data may simply not be available, particularly for therapeutic applications. As mentioned earlier, cross-reactivity is a substantial problem that plagues the development of GPCR antibodies. MULTISCREENTM uses flowcytometry and cell-based functional assays to screen for binding against a variety of GPCRs. These panels of GPCRs are customized to reflect the most related or liability targets for each specific antibody or therapeutic area. For all receptors that demonstrate antibody binding, dose-response curves can be generated.
For instance, Multispan employed a panel of 43 GPCRs including 5 other chemokine receptors to profile the binding of a commercially available antibody targeting CCR5. This assay was conducted under non-permeabilized conditions to limit staining to surface receptors in the extracellular domains. Binding, as measured by the fluorescent intensity, was normalized for receptor expression of each GPCR to ensure binding – not differential receptor expression – was being compared. As you can see below, the CCR5 antibody showed significant cross-reactivity with other assayed receptors.
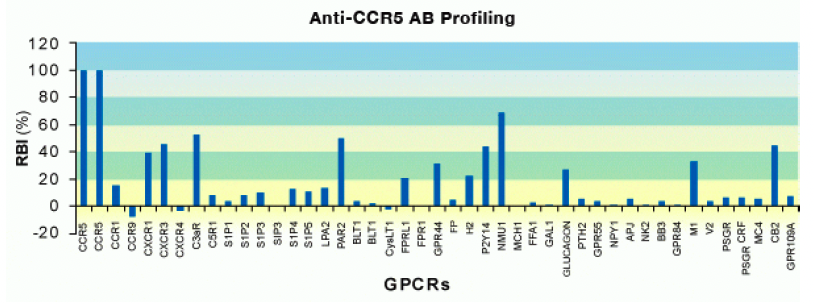
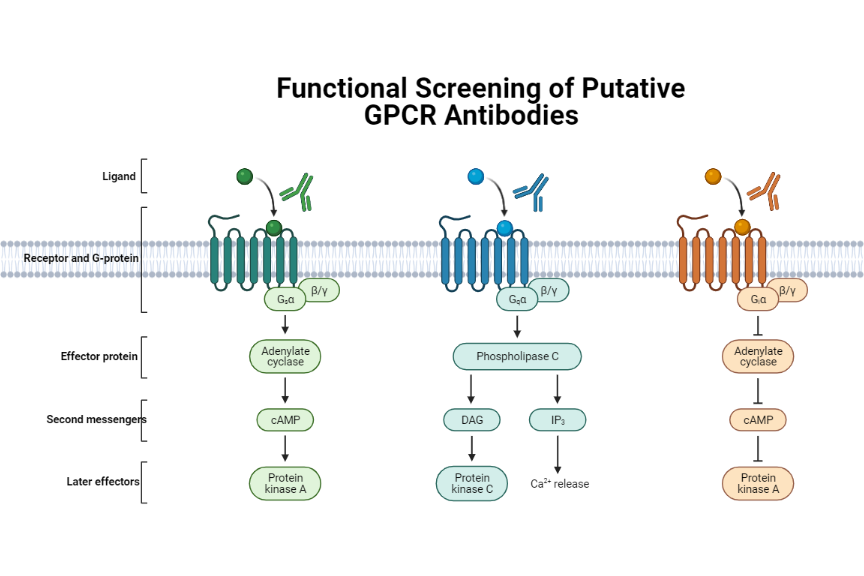
Though the FACS-based panels are useful to measure off-target binding, functional assays to test cross-reactivity are far more powerful. This is where the strengths of Multispan’s expertise in GPCR functional assays can be harnessed for antibody validation. MULTISCREENTM 32-GPCR safety functional panel can be employed to ensure the safety of a lead compound whereas the extensive MULTISCREENTM 231-GPCR cell-based assay panel that includes 66 receptor subfamilies ensures target specificity. To warrant assay consistency, each GPCR target in the MULTISCREENTM assay panels is expressed in a single, stable clonal cell line that has been carefully characterized. This panel utilizes receptor-dependent, G protein-coupled cellular secondary messenger readouts (e.g., calcium, cAMP). Multispan functional assays are physiologically relevant, high-throughput options for screening antibodies for research and therapeutic purposes.
References
- Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017 Dec;16(12):829-842. doi: 10.1038/nrd.2017.178. Epub 2017 Oct 27. PMID: 29075003; PMCID: PMC6882681.
- Dolgin, E. First GPCR-directed antibody passes approval milestone. Nat Rev Drug Discov 17, 457–459 (2018). https://doi.org/10.1038/nrd.2018.103
- O’Hayre M, Vázquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, Gutkind JS. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer. 2013 Jun;13(6):412-24. doi: 10.1038/nrc3521. Epub 2013 May 3. PMID: 23640210; PMCID: PMC4068741.
- Orbán E, Proverbio D, Haberstock S, Dötsch V, Bernhard F. Cell-free expression of G-protein-coupled receptors. Methods Mol Biol. 2015;1261:171-95. doi: 10.1007/978-1-4939-2230-7_10. PMID: 25502200.
- Takeda, H., Ogasawara, T., Ozawa, T. et al. Production of monoclonal antibodies against GPCR using cell-free synthesized GPCR antigen and biotinylated liposome-based interaction assay. Sci Rep 5, 11333 (2015). https://doi.org/10.1038/srep11333
- Heydenreich FM, Vuckovic Z, Matkovic M and Veprintsev DB (2015) Stabilization of G protein-coupled receptors by point mutations. Front. Pharmacol. 6:82. doi: 10.3389/fphar.2015.00082
- Talmont F, Moulédous L, Boué J, Mollereau C, Dietrich G. Denatured G-protein coupled receptors as immunogens to generate highly specific antibodies. PLoS One. 2012;7(9):e46348. doi: 10.1371/journal.pone.0046348. Epub 2012 Sep 27. PMID: 23029489; PMCID: PMC3459905.
- Mirzabekov T, Kontos H, Farzan M, Marasco W, Sodroski J. Paramagnetic proteoliposomes containing a pure, native, and oriented seven-transmembrane segment protein, CCR5. Nat Biotechnol. 2000 Jun;18(6):649-54. doi: 10.1038/76501. PMID: 10835604.
- Yan H, Gu W, Yang J, Bi V, Shen Y, Lee E, Winters KA, Komorowski R, Zhang C, Patel JJ, Caughey D, Elliott GS, Lau YY, Wang J, Li YS, Boone T, Lindberg RA, Hu S, Véniant MM. Fully human monoclonal antibodies antagonizing the glucagon receptor improve glucose homeostasis in mice and monkeys. J Pharmacol Exp Ther. 2009 Apr;329(1):102-11. doi: 10.1124/jpet.108.147009. Epub 2009 Jan 7. PMID: 19129372.
- Ferrara, F., Erasmus, M.F., D’Angelo, S. et al. A pandemic-enabled comparison of discovery platforms demonstrates a naïve antibody library can match the best immune-sourced antibodies. Nat Commun 13, 462 (2022). https://doi.org/10.1038/s41467-021-27799-z
- Alfaleh MA, Alsaab HO, Mahmoud AB, Alkayyal AA, Jones ML, Mahler SM and Hashem AM (2020) Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Front. Immunol. 11:1986. doi: 10.3389/fimmu.2020.01986
- Moore, P., Clayton, J. To affinity and beyond. Nature 426, 725–727 (2003). https://doi.org/10.1038/426725a
- Pedrioli A, Oxenius A. Single B cell technologies for monoclonal antibody discovery. Trends Immunol. 2021 Dec;42(12):1143-1158. doi: 10.1016/j.it.2021.10.008. Epub 2021 Nov 4. PMID: 34743921.
- Baker, M. Reproducibility crisis: Blame it on the antibodies. Nature 521, 274–276 (2015). https://doi.org/10.1038/521274a
Download PDF

Recent Comments