G protein-coupled receptors (GPCRs) are highly dynamic membrane proteins representing the most leading family of validated drug targets in biomedicine. More than 800 genes comprise this receptor family that are involved in numerous physiological processes and govern every aspect of how an organism functions (1). Despite significant progress in understanding the molecular mechanism of GPCR activation, the structural and functional aspects of GPCRs (especially orphan receptors) as a whole is not fully understood. Filling this gap in knowledge is absolutely necessary to understand how GPCRs bind to numerous ligands and transmit diverse signals to elicit physiological functions.
What is mutagenesis?
Mutagenesis is the process in which mutations are purposely introduced into the receptor. With recent advent in polymerase chain reaction technology, random or targeted (site-directed) mutagenesis is routinely used for the study of GPCR structure-function relationships (2). Random mutagenesis introduces mutations in a random fashion and thus is non-specific while site-directed mutagenesis refers to single mutation as well as double or multiple combined mutations including single-residue mutation, deletion, and insertion (2).
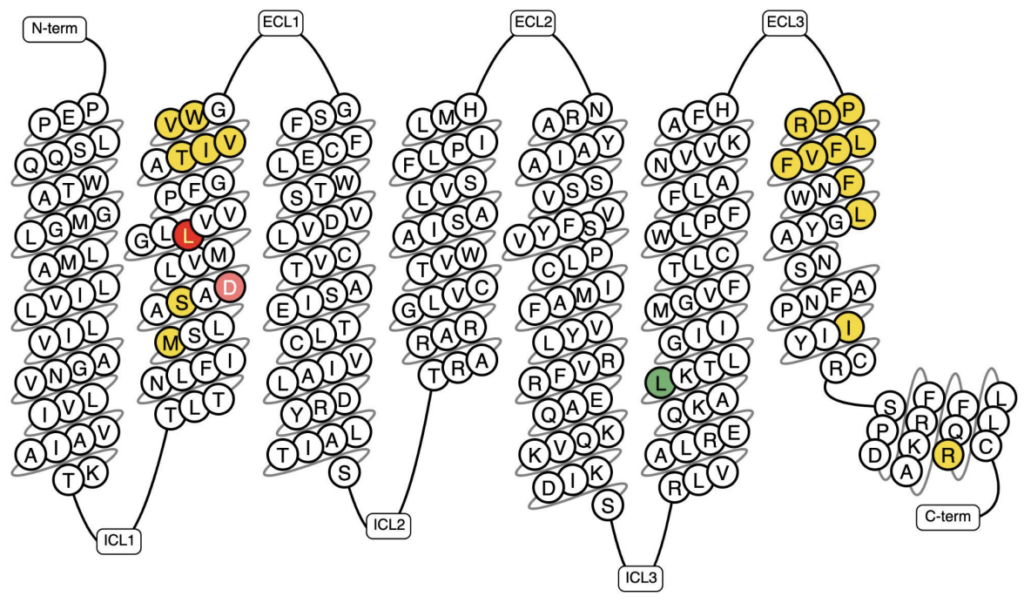
Figure 1: Snake plot of the β1-adrenergic receptor with documented known in-vitro mutants highlighted. Invitro Mutant Data: Increased binding/potency: >10-fold; Reduced binding/potency: >5-fold, >10-fold; No/low effect (<5-fold); and N/A (18, 19).
How can mutagenesis help probe GPCR structure/function relationships, thus accelerating drug discovery for GPCR targets?
Starting with crystal structure determination and accelerated by cryo-electron microscopy (cryo-EM), technology has finally enabled rational GPCR drug discovery programs such as structure-guided drug design (3). Although their structural characterization has been impeded by low expression yields and receptor instability in detergents, making such efforts extremely challenging, if not impossible (4), progress is being made. In fact, this is the motivation behind developing stabilizing mutations to characterize the structures of GPCRs that impart native-like pharmacological and biophysical properties. High-resolution receptor dynamics that locks the receptor at a particular conformation obtained by mutation(s) at precise locations in the receptor can offer additional opportunities for drug design
Apart from crystallography and cryo-EM, mutagenesis strategies are also used for nuclear magnetic resonance (NMR) spectroscopy, surface plasmon resonance (SPR), native mass spectrometry, and fluorescence anisotropy measurements, all of which are important to study the complex aspects of the receptor dynamics (5). Another critical aspect of mutagenesis research is the study of naturally occurring mutations including single-nucleotide polymorphisms (SNPs) in the population and their correlations with health and diseases (6) This can validate the receptors as targets and link the signaling pathways to potential therapy.
How do receptor mutants contribute to understanding GPCR dynamics?
1. Stabilization of GPCR by mutagenesis for cryo-EM applications
GPCRs exist in multiple inactive and active conformational states that are in dynamic equilibrium. Stabilization by mutagenesis can help trap the receptor in a desired conformation. One of these is alanine scanning, an extensive mutagenesis approach, aimed at finding single stabilizing mutations where the residues in the receptor are systematically substituted for alanine one by one (7). This can be exemplified by the conformational thermostabilization of turkey β1-adrenergic receptor (C116L) in the antagonist-bound state which was critical for its subsequent structure determination at 2.7 Å resolution (8). Other mutant scanning approaches include lysine scanning, which has been used to investigate the ligand binding region of nicotinic acetylcholine receptor delimiting a hydrophobic core (9). Moreover, cysteine mutations have also been extensively used to capture different conformational states of the receptor essential for signal transduction. For example, engineering of disulphide bonds in the parathyroid hormone 1 receptor between TM6 and TM7 provided insights into the early transduction mechanism of the receptor activation (10). Thus, mutagenesis analyses combined with structural data have proven to be powerful tools to probe ligand-receptor and receptor-receptor interfaces.
2. Thermostabilization of GPCR for biophysical applications
Beyond cryo-EM, biophysical characterization of GPCRs provide complementary sources of structural information about the receptor activation and its signaling. However, the loss of native structure upon solubilization in detergent and low thermostability of GPCRs have always presented major challenges for their biophysical, and biochemical characterization (5). Conformationally thermostabilized receptors obtained by mutagenesis allows the receptor to function at high temperatures for a prolonged period of time, which is usually not possible with native GPCRs (11). Also, thermostabilized GPCRs are well suited for methods requiring long half-life in detergent such as SPR or NMR. A representative example is adenosine2A and 𝛃1-adrenergic receptor stabilized by alanine scan for extensive SPR screenings, enabling biophysical mapping of the ligand binding site and thus fragment-based drug discovery (12).
3. Studying the mechanism of action underlying GPCR pharmacology
Mutagenesis approaches can also be utilized to study the pharmacological properties of GPCRs. Introduction of mutations in the binding domain of the receptor have been used to identify fundamentals of GPCR function to guide research in molecular pharmacology. Understanding the mechanisms of receptor desensitization and internalization events requires determining which domains and specific amino acid residues are involved upon agonist stimulation. For example, point- and cluster-mutants with alanine substituted for serine or threonine residues located in the C-termini of the opioid receptor was constructed to understand the mechanism underlying the DAMGO-induced µ opioid receptor phosphorylation and desensitization (13).
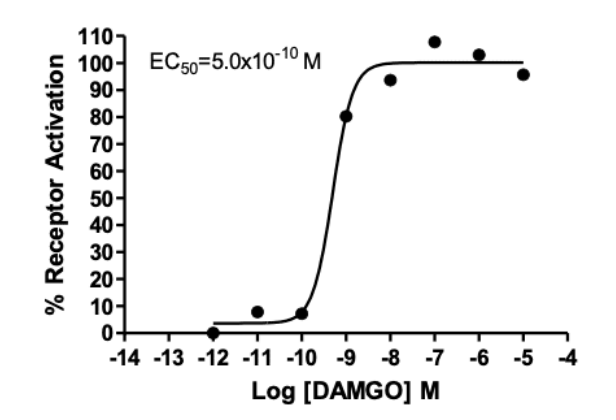
Figure 2: Dose-dependent stimulation of pERK level upon treatment with ligand, monitored with FlexStation. Source: Multispan’s µ opioid multiscreen stable cell lines
4. Mutant GPCRs as a cause of human diseases
Mutations in GPCRs can significantly affect their physiological function which often lead to gain or loss of function phenotypes. To date, over 60 human monogenic diseases are directly caused by approximately 2.3K mutations in 55 GPCR genes (14). In their review, Schöneberg & Liebscher also report that approximately 14 diseases are cause by gain of function mutations in GPCRs and that to date 7 of these mutated GPCRs are orphan receptors that contribute to almost 10% of known disease caused by mutant receptors (14, 15). Understanding the role of genetic variants in GPCRs as well as the precise signaling pathways altered by these mutations is more important than ever for elucidating disease pathogenesis mechanisms (14,16). Genetic variants/SNPs identified in GPCRs can influence receptor expression, targeting, function, and receptor turnover; as well as the ability of the receptor to recognize and respond to ligands (14, 16). Better characterizing genetic variants and SNPs is a powerful approach to designing strategies to rescue and improve their druggability.
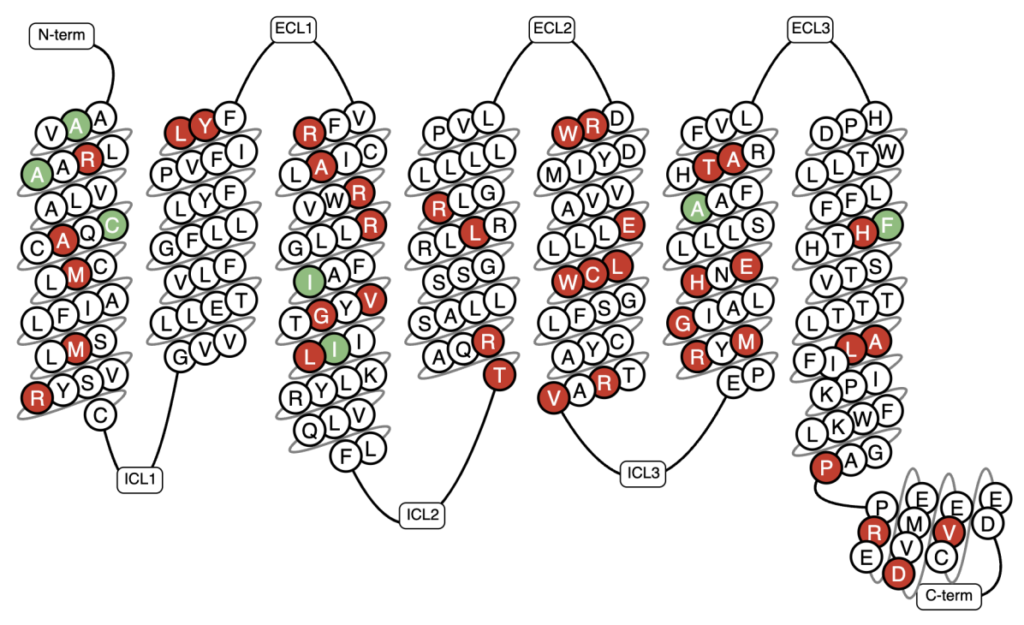
Figure 3: Snake plot of the orphan receptor GPR179 with its known genetic variations highlighted (18, 19). Mutations in GPR179 cause congenital stationary night blindness discussed in Ref. 14. Invitro Mutant Data: Increased binding/potency: >5-fold & Reduced binding/potency: >10-fold.
Conclusions
To summarize, in spite of all the technological and scientific advances in the field, there is still more work to be done to fully exploit the druggability of GPCRs to their fullest potential (17).
Multispan’s mission is to support scientists in both industry and academia. We provide GPCR focused high quality, reliable assay service, ready-to-use tools and kits in addition to being a leader in cell engineering to help develop better medicines one assay at a time. We carry a large variety of ready-to-use tools for GPCR structure/function studies including but not limited to GPCRs cited in this review for which ready-to-use stable cell lines are available can be explored: Adenosine 2AR, β1-adrenergic, µ opioid receptor.
Get in touch today and let us help you find cures one assay at a time.
References
- Kapolka, N. J., Taghon, G. J., Rowe, J. B., Morgan, W. M., Enten, J. F., Lambert, N. A., & Isom, D. G. (2020). DCyFIR: a high-throughput CRISPR platform for multiplexed G protein-coupled receptor profiling and ligand discovery. Proceedings of the National Academy of Sciences, 117(23), 13117-13126.
- Conner, A. C., Barwell, J., Poyner, D. R., & Wheatley, M. (2011). The use of site-directed mutagenesis to study GPCRs. In Receptor Signal Transduction Protocols (pp. 85-98). Humana Press, Totowa, NJ.
- Jacobson, K. A. (2015). New paradigms in GPCR drug discovery. Biochemical pharmacology, 98(4), 541-555.
- Waltenspühl, Y., Ehrenmann, J., Klenk, C., & Plückthun, A. (2021). Engineering of Challenging G Protein-Coupled Receptors for Structure Determination and Biophysical Studies. Molecules, 26(5), 1465.
- Heydenreich, F. M., Vuckovic, Z., Matkovic, M., & Veprintsev, D. B. (2015). Stabilization of G protein-coupled receptors by point mutations. Frontiers in pharmacology, 6, 82.
- Insel, P. A., Tang, C. M., Hahntow, I., & Michel, M. C. (2007). Impact of GPCRs in clinical medicine: monogenic diseases, genetic variants and drug targets. Biochimica et Biophysica Acta (BBA)-Biomembranes, 1768(4), 994-1005.
- Horovitz, A., Matthews, J. M., & Fersht, A. R. (1992). α-Helix stability in proteins: II. Factors that influence stability at an internal position. Journal of molecular biology, 227(2), 560-568.
- Serrano-Vega, M. J., Magnani, F., Shibata, Y., & Tate, C. G. (2008). Conformational thermostabilization of the β1-adrenergic receptor in a detergent-resistant form. Proceedings of the National Academy of Sciences, 105(3), 877-882.
- Sine, S. M., Wang, H. L., & Bren, N. (2002). Lysine scanning mutagenesis delineates structural model of the nicotinic receptor ligand binding domain. Journal of Biological Chemistry, 277(32), 29210-29223.
- Thomas, B. E., Woznica, I., Mierke, D. F., Wittelsberger, A., & Rosenblatt, M. (2008). Conformational changes in the parathyroid hormone receptor associated with activation by agonist. Molecular Endocrinology, 22(5), 1154-1162.
- Sarkar, C. A., Dodevski, I., Kenig, M., Dudli, S., Mohr, A., Hermans, E., & Plückthun, A. (2008). Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proceedings of the National Academy of Sciences, 105(39), 14808-14813.
- Popov, P., Peng, Y., Shen, L., Stevens, R. C., Cherezov, V., Liu, Z. J., & Katritch, V. (2018). Computational design of thermostabilizing point mutations for G protein-coupled receptors. Elife, 7, e34729.
- Deng, H. B., Yu, Y., Pak, Y., O’Dowd, B. F., George, S. R., Surratt, C. K., … & Wang, J. B. (2000). Role for the C-terminus in agonist-induced μ opioid receptor phosphorylation and desensitization. Biochemistry, 39(18), 5492-5499.
- Schöneberg & Liebscher (2021) Mutations in G Protein–Coupled Receptors:Mechanisms, Pathophysiology and Potential Therapeutic Approaches. Pharmacol Rev. 2021 Jan;73(1):89-119.
- Hauser AS, Gloriam DE, Bräuner-Osborne H, and Foster SR (2020) Novel approaches leading towards peptide GPCR de-orphanisation. Br J Pharmacol 177:961–968.
- Balasubramanian, S., Xia, Y., Freinkman, E., & Gerstein, M. (2005). Sequence variation in G-protein-coupled receptors: analysis of single nucleotide polymorphisms. Nucleic acids research, 33(5), 1710-1721.
- Yang, D., Zhou, Q., Labroska, V., Qin, S., Darbalaei, S., Wu, Y., … & Wang, M. W. (2021). G protein-coupled receptors: Structure-and function-based drug discovery. Signal transduction and targeted therapy, 6(1), 1-27.
- GPCRdb in 2021: integrating GPCR sequence, structure and function. Kooistra AJ, Mordalski S, Pandy-Szekeres G, Esguerra M, Mamyrbekov A, Munk C, Keseru GM, Gloriam DE Nucleic acids research, 2021, 49:D335-D343
- GPCRdb: an information system for G protein-coupled receptors. Isberg V, Mordalski S, Munk C, Rataj K, Harpsoe K, Hauser AS, Vroling B, Bojarski AJ, Vriend G, Gloriam DE. Nucleic acids research, 2016, 44:D356-64
Download PDF

Recent Comments